
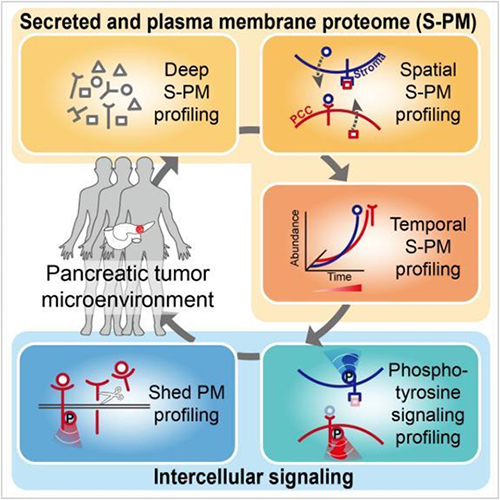
Figure Schematic diagram of signal transduction network between cells in
tumor microenvironment of pancreatic cancer analyzed by TMEPro
Under the support of the National Natural Science Foundation of China
(Grant No.: 22125403, 92253304), Professor Tian Ruijun's team of Southern
University of Science and Technology developed the multi-dimensional clinical
functional proteomics analysis strategy TMEPro, and cooperated with Professor
Qin Renyi's team of Tongji Hospital of Huazhong University of Science and
Technology, Gao Dong's research team of the Center for Excellence and Innovation
in Molecular Cell Science of the Chinese Academy of Sciences, and Dr. Yu Shi of
the Salk Institute of the United States to achieve systematic biomedical
application research on pancreatic cancer. The research achievement is entitled
"Clinical functional proteomics of intercellular signaling in pancreatic cancer"
and published in Nature on November 13, 2024. The link of the paper is as
follows: https://www.nature.com/articles/s41586-024-08225-y .
Pancreatic ductal adenocarcinoma (PDAC) is one of the deadliest cancers,
with a 5-year survival rate of less than 10%. It is very important to develop
reliable biomarkers for early diagnosis and effective targeted drugs for the
treatment of pancreatic cancer. The tumor microenvironment (TME) is the most
representative feature of cancer, and PDAC TME is atypical to be rich in a large
number of non malignant stromal cells and extracellular matrix, which interact
closely with tumor cells and promote their proliferation, metastasis, and drug
resistance. Proteomic analysis based on mass spectrometry can systematically
explore the signal transduction process between PDAC cells, but current
proteomic research is mostly conducted at the cellular level or using block
tumor samples, which can only obtain average analysis results or are difficult
to fully reveal the true functional proteomic characteristics in clinical
samples.
In response to this issue, the aforementioned team has developed the
clinical functional proteomics analysis strategy TMEPro, which starts from the
underlying principles, wet experiments, and dry experiments, and uses various
chemical measurement and chemical biology strategies to analyze the functional
proteomic characteristics in tumor samples from multiple dimensions,
systematically solving the technical difficulties mentioned above. By developing
TMEPro, the team directly analyzed the spatiotemporal dynamic proteome from
trace PDAC clinical samples from five dimensions, overcoming the limitations of
traditional research methods such as block tissue samples or cultured cell
lines. Further combined with multidimensional bioinformatics analysis, a
comprehensive intercellular signal transduction network map was constructed,
revealing the functional secretion of ligand receptor membrane protein
interactions between cancer cells and stromal cells. This study found the
splicing of the receptor tyrosine kinase AXL outer membrane in cancer cells and
the cross signal axis between cancer cells and stromal cells, providing a useful
functional proteome database and potential molecular typing and targeted
treatment schemes for accurate diagnosis and treatment of pancreatic cancer.