
Figure 2 Modular modeling and intelligent design of enhancer based on
TFBU
With the support of the National Natural Science Foundation of China
projects (approval numbers: 62250007, 62225307) and other grants, Professor Wang
Xiaowei's team from the Department of Automation at Tsinghua University has made
progress in the intelligent design of gene regulatory sequences in synthetic
biology. The series of research results have been published in two consecutive
papers: (1) "Systematic representation and optimization enable the inverse
design of cross species regulatory sequences in bacteria" was published in the
journal Nature Communications on February 19, 2025. Paper link:
https://doi.org/10.1038/s41467-025-57031-1 (2) The title "Modeling and designing
enhancers by introducing and utilizing transcription factor binding units" was
published on February 8, 2025 in the journal Nature Communications. Paper link:
https://www.nature.com/articles/s41467-025-56749-2 .
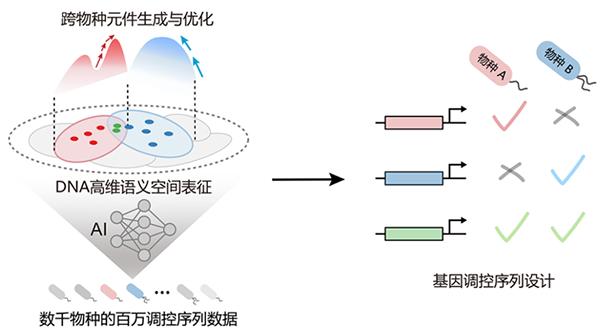
In response to the bottleneck problem of poor cross host adaptation of gene
circuits in biomanufacturing, the research team characterized functional
regulatory sequences as conditional probability distributions in the DNA
sequence space from the perspective of information encoding, and found that
cross species regulatory rules are hidden in the overlapping regions of
different species' conditional probability distributions; By integrating
millions of functional sequence data from thousands of species, a
high-dimensional semantic representation space and intelligent generation model
for DNA across species boundaries were constructed, breaking through the species
barrier of natural components. Experimental results showed that the model
achieved a 93.3% accuracy in cross host sequence adaptation in Escherichia coli
and Pseudomonas aeruginosa (Figure 1). In addition, a new transcription factor
binding unit (TFBU) model has been proposed to address the challenge of
quantitative modeling of gene enhancers in mammalian cells; This model models
the core binding sites of transcription factors and their surrounding
environmental sequences as a functional whole, breaking through the limitations
of traditional methods that only focus on local combinations of binding sites
and ignore the global effects of sequence context. It successfully quantifies
the impact of environmental sequences on transcription factor binding and
enhancer activity, providing a new tool for the development of novel therapies
such as gene therapy (Figure 2).
The series of studies combines intelligent model driven digital evolution
with active learning driven synthetic biology experiments, significantly
improving the design efficiency of synthetic biology sequences through
collaborative exploration and closed-loop iterative optimization of the "virtual
world" and "material world".