
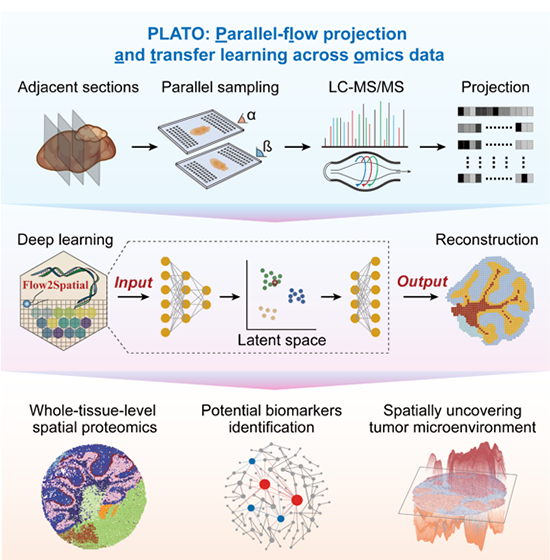
Schematic diagram of high-resolution spatial proteome detection and
analysis process at the level of whole tissue slices
Supported by the National Natural Science Foundation of China (Grant No.:
32025009, 32130020, 32400533, 32300538), Zhao Fangqing and Ji Peifeng from the
Institute of Zoology of the Chinese Academy of Sciences have made progress in
the field of artificial intelligence driven space proteomics technology. The
research results are entitled "High resolution spatially resolved proteins of
complex tissues based on microfluidics and transfer learning" and published
online in Cell on January 23, 2025) Magazine, paper link:
https://www.cell.com/cell/fulltext/S0092-8674 (24)01436-3#sec-5。
Space omics technology has become an important tool for analyzing tissue
heterogeneity and complex cellular interaction mechanisms. Especially, spatial
transcriptomics technology has shown great potential in embryonic development,
neuroscience, and disease mechanism research. However, existing spatial
proteomics techniques are limited in their application in complex tissue
research due to factors such as mass spectrometry detection flux and high cost,
making it difficult to meet the requirements of high-resolution and large-area
tissue analysis.
The research team proposed PLATO, an artificial intelligence driven spatial
proteomics measurement and analysis technology framework, which integrates deep
learning algorithms with microfluidic technology to achieve high-resolution
spatial proteome detection at the tissue slice level. Firstly, this study
combined microfluidic technology to develop a high-throughput, low-cost parallel
in-situ sampling platform that can achieve flexible and accurate sampling in the
resolution range of 25 to 100 micrometers. Secondly, this study uses artificial
intelligence algorithms to restore the spatial position information of proteins,
breaking through the limitations of traditional mass spectrometry technology in
obtaining spatial information and significantly improving the coverage and
resolution of spatial proteomics. The research team used this technology to
analyze the spatial distribution of proteins with high resolution in mouse brain
tissue, intestinal villi, breast cancer and other complex tissues, further
verifying the huge potential of this method in different application scenarios
and research directions.
This study deeply integrates artificial intelligence algorithms,
microfluidics, and mass spectrometry technologies, achieving important
breakthroughs and technological iteration innovations in spatial omics
technology. It provides strong support for revealing protein dynamic
distribution and exploring the molecular mechanisms of complex biological
processes, and is expected to play an important role in disease diagnosis,
precision medicine, and agricultural biotechnology.